Let's say you have a whole lot of csv files in your working directory. By some convenient act of divine grace they also happen to have the same column structures. Reading them into R can be a slow matter as a new R user may try to write out the name of every file, assign it to a variable and then rbind() it all together later on. A slightly more experienced user might choose to automate it a bit by using list.files() with a for loop to iterate through every csv file in the directory. A yet more advanced user could figure out via much cursing and pain how to do this using the apply() family of functions, which may actually be the quickest way to do this. For myself, I like to take headache-saving shortcuts when possible and still maintain some semblance of code efficiency, so naturally I use the eponymous plyr package for this task.
Just for example, let's make a bunch of fake csv files that will all have the same structure.
for(i in 1:100) {
df <- data.frame(x=rnorm(100), z=runif(100))
write.csv(df, sprintf('file%d.csv', i))
}
Then we can write up a convenience function to load plyr, find all the csv's, and define a function that we will run based on the user-entered arguments. Keep in mind you're going to want to pass the output of this function into a variable.
read.directory <- function(dir, stringsAsFactors=F, keepMeta=F) {
require(plyr)
files <- list.files(dir, pattern='\\.csv', ignore.case=T)
toExec <- "mdply(files, read.csv, stringsAsFactors=stringsAsFactors)"
if(!keepMeta) {
toExec <- paste0(toExec, "[, -c(1, 2)]")
}
return(eval(parse(text=toExec)))
}
You're probably wondering about the "keepMeta" argument. When you run mdply() without adding the column subset to the end of it you end up with two extra columns: one for the index number of the file it came from in list.files(), the second being the actual row number that record resided in within that file. I find that info to be unnecessary most of the time, hence the default of "keepMeta=F" re-writing the function so that it excludes the offending columns.
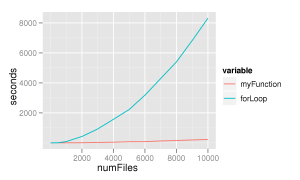
As a side note about this, I ran my function against a for loop to see how well it performed. I believe the results are quite clear and provide yet another piece of evidence as to why you should vectorize your code whenever possible. Additionally, the *ply functions in plyr support multi-core execution which could cause the function to execute even quicker. This is an area I'll have to research perhaps for a "Part 2". The for loop used is after the image. In both cases, they were simply dropped into system.time() and the "User" time recorded. It was done for 10, 100, 500, 1000, 2000, ..., 10000 files.
for (i in list.files('.', pattern='\\.csv', ignore.case=T)) {
myData <- rbind(try(myData, TRUE), read.csv(i, stringsAsFactors=FALSE))
}
One additional thing I learned is that when testing for loops you really should write a script to automate it. It took more time for all of the for loops to finish than it did to come up with the idea for this post, write the function, test the function, get badgered my co-blogger for taking forever to get this post up, and write all the portions of the blogpost not relating to testing the for loop.
Also, I apologize for the weird formatting. I was hoping to figure out how to get WordPress to respect my code indentation, but it doesn't seem to agree with me on that. Hopefully you can forgive this fact as a first post while we try to figure that bit out.